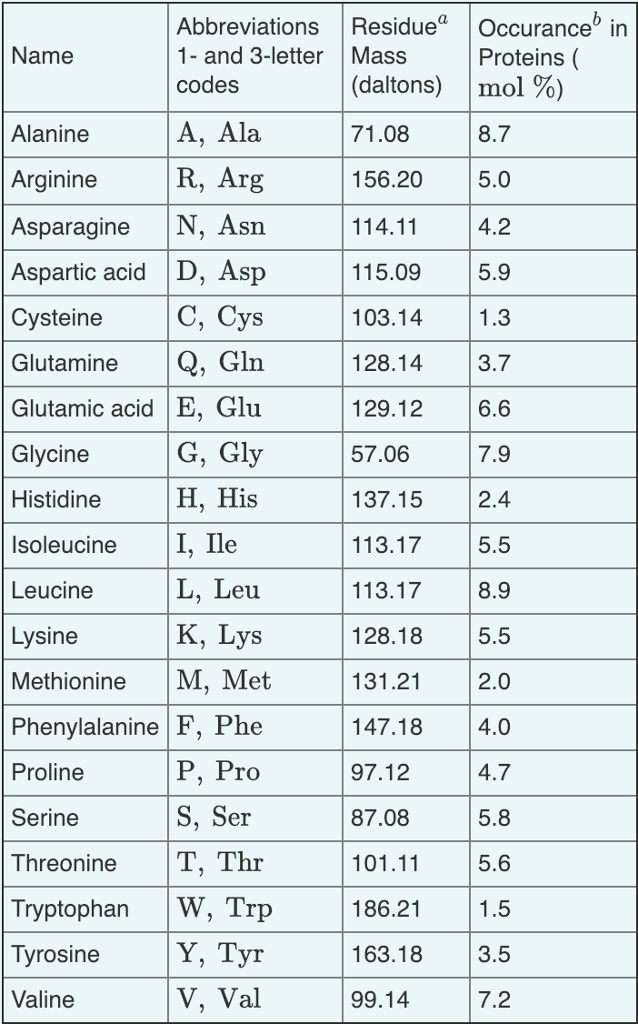
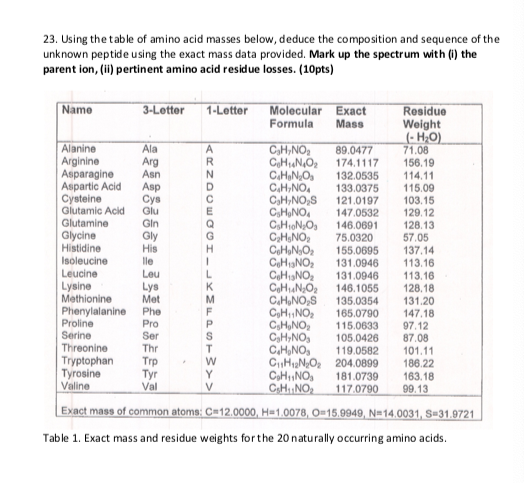
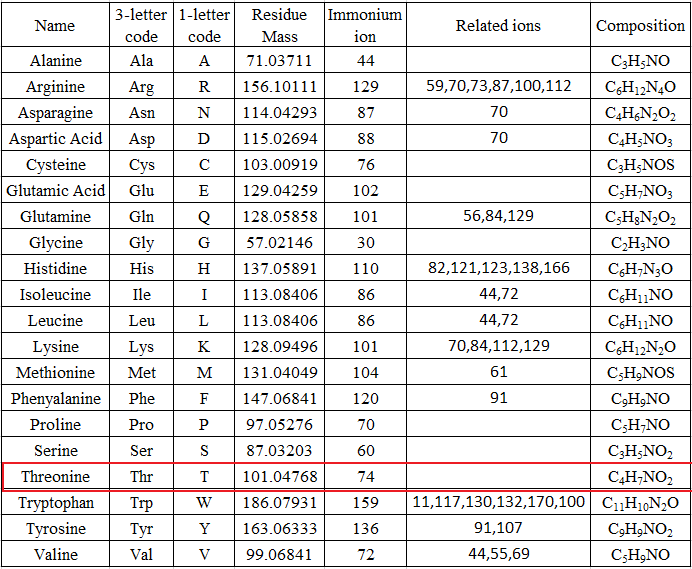
Table 1 from Characterization of Ni(II) complexes of Schiff bases of amino acids and (S)-N-(2-benzoylphenyl)-1-benzylpyrrolidine-2-carboxamide using ion trap and QqTOF electrospray ionization tandem mass spectrometry. | Semantic Scholar

Gut amino acid absorption in humans: Concepts and relevance for postprandial metabolism - Clinical Nutrition Open Science

Characterization of chiral amino acids from different milk origins using ultra-performance liquid chromatography coupled to ion-mobility mass spectrometry | Scientific Reports

of possible combinations of amino acids based on molar mass±0.2 mass... | Download Scientific Diagram
Evaluation of open search methods based on theoretical mass spectra comparison | BMC Bioinformatics | Full Text

Expanded Cellular Amino Acid Pools Containing Phosphoserine, Phosphothreonine, and Phosphotyrosine | ACS Chemical Biology

Table 1 from Mass-based classification (MBC) of peptides: Highly accurate precursor ion mass values can be used to directly recognize peptide phosphorylation | Semantic Scholar

Evaluation of NHS-Acetate and DEPC labelling for determination of solvent accessible amino acid residues in protein complexes - ScienceDirect

Mono-dibase codon-amino acid" logic. Order of hits (from the Table 1)... | Download High-Resolution Scientific Diagram